PROJECT TITLE: EukRef: The 18S annotation initiative, 3rdWokshop
Acronym:
Scope: Global
Type: Research and Workshop
Timeframe: 4-9 November 2018
Principal Investigator: Dr. Javier del Campo
Address: Passeig Marítim de la Barceloneta 37-49, 08003 Barcelona, Catalonia
Tel/Fax: +34 616342889
E-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
Home page URL: http://emm.icm.csic.es/
Other key persons (name, title and institution):
Dr. Cédric Berney, UniEuk Taxonomic Coordinator, Station Biologique Roscoff, France
Objectives:
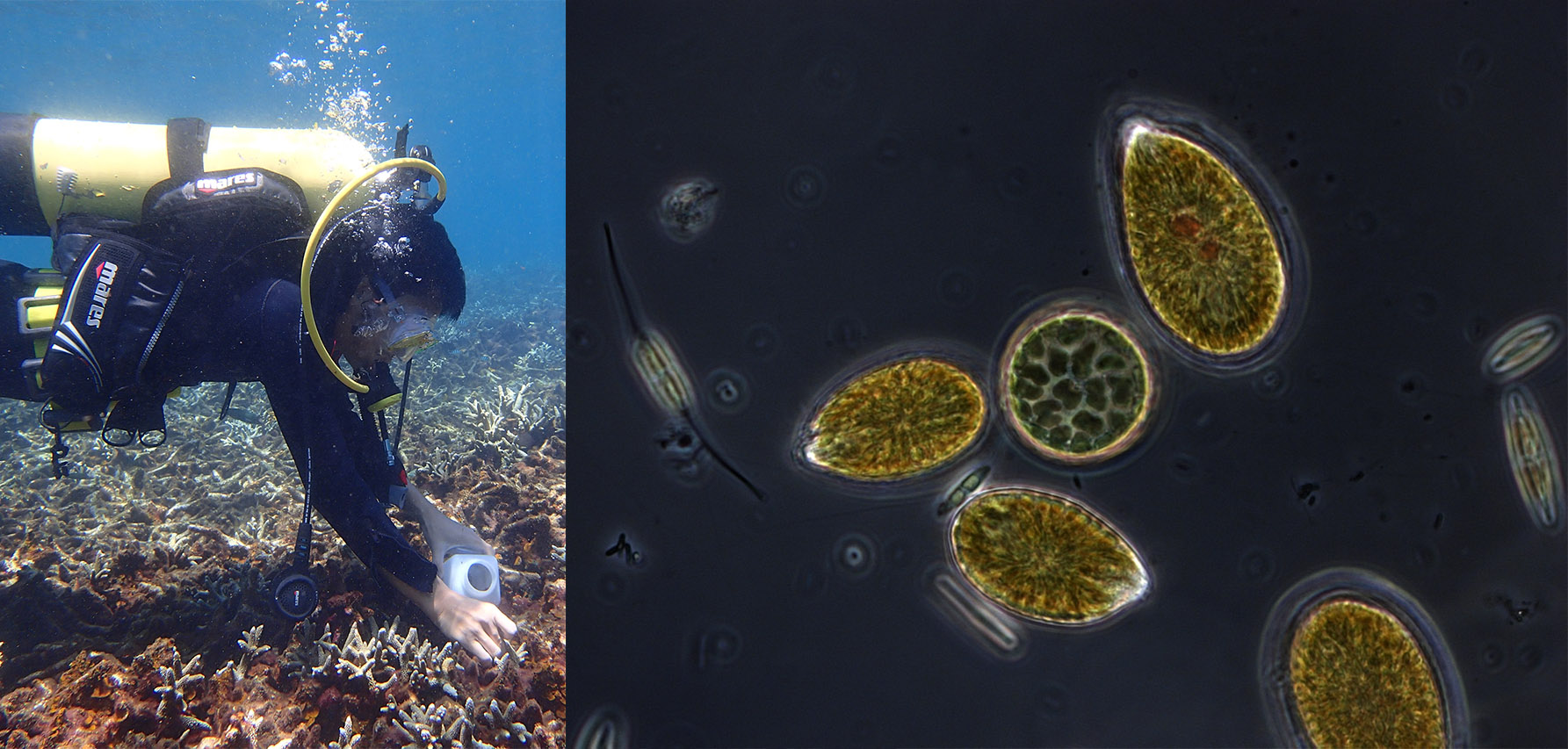
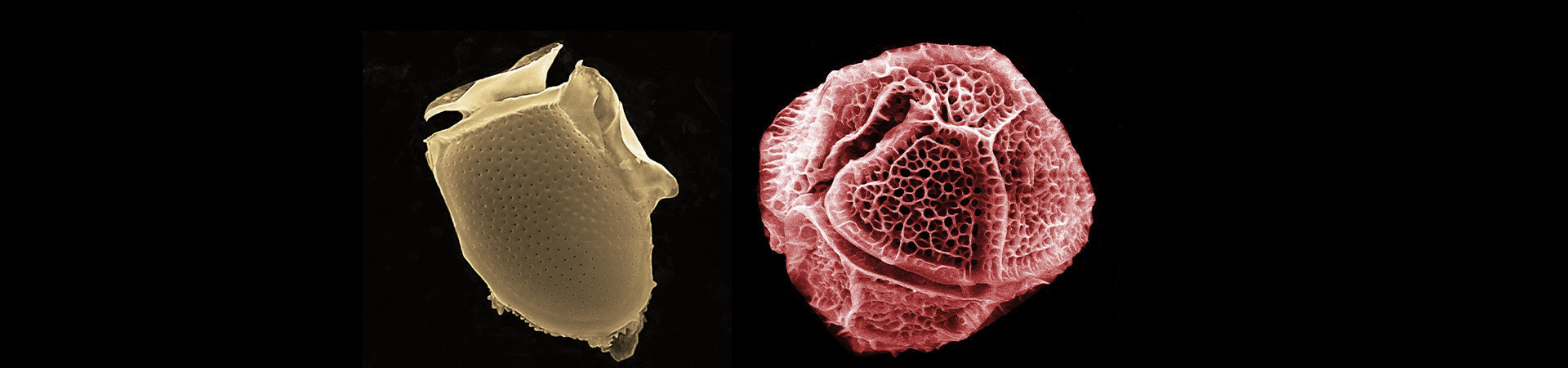
Environmental sequencing has greatly expanded our knowledge of micro-eukaryotic diversity and ecology by revealing previously unknown lineages and their distribution. However, the value of these data is critically dependent on the quality of the reference databases used to assign an identity to environmental sequences. Existing databases contain errors, and struggle to keep pace with rapidly changing eukaryotic taxonomy, the influx of novel diversity, and computational challenges related to assembling the high-quality alignments and trees needed for accurate characterization of lineage diversity.
UniEuk (www.unieuk.org) is an open, community-based and expert-driven international initiative to build a flexible, adaptive universal taxonomic framework for eukaryotes, focused primarily on protists. The UniEuk system comprises 3 complementary modules allowing direct community input:
- EukRef, a standardized, open-source bioinformatics pipeline that allows taxonomic curation of publicly available phylogenetic marker sequences (starting with 18S rDNA), generating homogeneous sets of curated, aligned sequences and phylogenetic trees.
- EukBank, a public repository of high-throughput metabarcoding datasets (starting with the V4 region of 18S rDNA) that allows monitoring of total eukaryotic diversity (e.g. saturation, phylogeny) across biomes, and identification of ecologically relevant new lineages.
- EukMap, a user-friendly representation of the taxonomic framework in the form of a publicly navigable tree, fully editable by registered users, where each node/taxon is associated with standardized features (name, contextual data, links to pictures and literature, etc.).
EukRef (eukref.org) is an ongoing community-driven initiative that addresses the above mentioned challenges by bringing together taxonomists with expertise spanning the eukaryotic tree of life and microbial ecologists that use environmental sequence data to develop reliable reference databases across the diversity of microbial eukaryotes. EukRef organizes and facilitates rigorous mining and annotation of sequence data by providing protocols, guidelines and tools. The EukRef pipeline and tools allow users interested in a particular group of microbial eukaryotes to retrieve all sequences belonging to that group from INSDC (GenBank, ENA or DBJD), to place those sequences in a phylogenetic tree, and to curate taxonomic and environmental information for the group. We provide guidelines to facilitate the process and to standardize taxonomic annotations. The final outputs of this process are 1) a reference tree and alignment, 2) a reference sequence database including taxonomic and environmental information, and 3) a list of putative chimeras and other artifactual sequences. These products will be useful for the broad community as they become publicly available (at eukref.org) and are shared with existing reference databases.
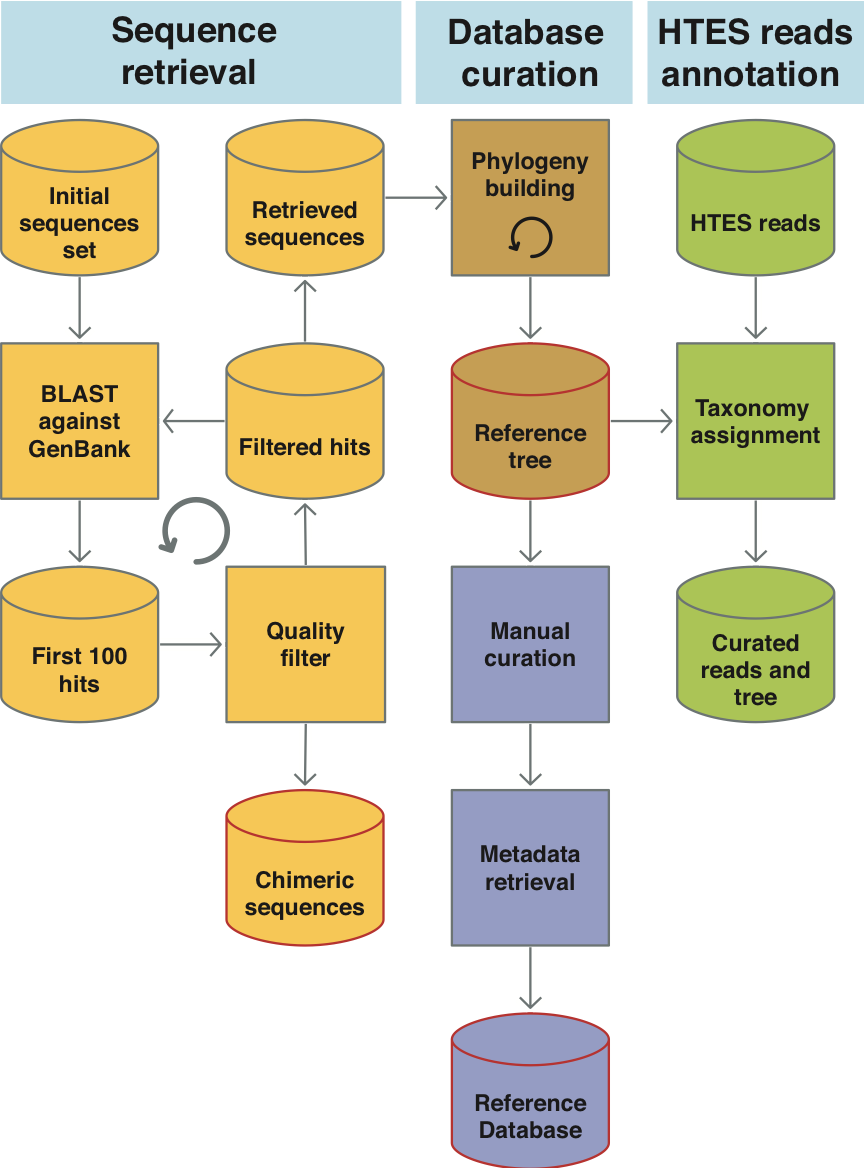 Implementation progress :On 2020, EukRef merged with Protist Ribosomal Reference Database (PR2), an eukaryotic barcoding database. Information on the future progresses can be found at the PR2 database web site.
Implementation progress :On 2020, EukRef merged with Protist Ribosomal Reference Database (PR2), an eukaryotic barcoding database. Information on the future progresses can be found at the PR2 database web site.